Theme 1 : Characterisation of enzymes involved in Bacillus subtilis RNA maturation and decay
The control of messenger RNA degradation is a highly efficient means of modulating gene expression. Our group studies the RNA maturation and degradation pathways of the Gram-positive soil bacterium, Bacillus subtilis. Our understanding of the pathways of RNA processing and turnover in B. subtilis has developed rapidly in the past few years. Of more than 35 bacterial RNases discovered in bacteria to date, the vast majority were identified in either E. coli or B. subtilis. Yet these organisms have only nine enzymes in common, and these are among the most highly conserved RNases in nature. Some enzymes that are essential in E. coli are completely absent in B. subtilis, and vice versa.
There are currently two major pathways known for mRNA turnover in B. subtilis. In the first, the mRNA is first cleaved by an endonuclease, in most cases by the membrane-bound RNase Y, the functional analog of E. coli RNase E. The fragments generated by endonucleolytic cleavage are then degraded in the 3’-5’ direction, principally by PNPase, and in the 5’-3’ direction by the RNase J1/J2 complex. Remarkably, E. coli does not posses 5’-3’ exoribonuclease activity. In the second pathway, an RNA pyrophosphohydralase deprotects primary transcripts by removing their 5’ triphosphate group. The 5’-monophosphorylated RNA is then degraded to completion by the 5’-3’ exoribonuclease activity of RNase J1/J2 This pathway is very similar to the eukaryotic mRNA degradation pathway catalysed by decapping enzymes and Xrn1.
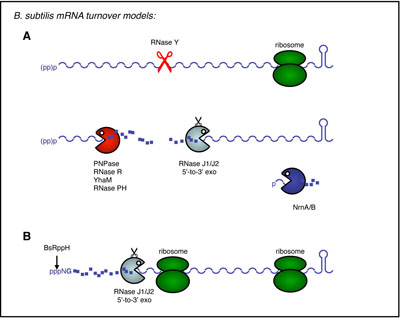
The group’s latest findings have provided insight into the degradation of B. subtilis mRNAs by:
